Our vision is to expand precision medicine from the current gene-centric process to one that considers the complexity and dynamics of proteins.
Our research is focused on the origin, mechanistic role, and clinical application of oncoproteoforms (proteins modified by cancer pathologies). We aim to improve precision therapies for young people (children to young adults) with cancer, based on the patient’s individual physiology rather than only their cancer’s genetics.
![]() Personalized oncology & molecular pathology in BRAvE & PROFYLE:
Personalized oncology & molecular pathology in BRAvE & PROFYLE:
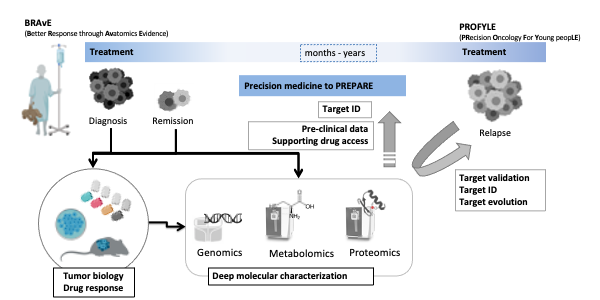
To advance molecular pathology and pre-clinical studies for kids with cancer we co-founded the Better Response through Avatomics Evidence (BRAvE) precision medicine program at BCCH and co-lead the proteomics node in the national Precision Oncology for Young People (PROFYLE) initiative.
We have established data-independent acquisition (DIA) based deep quantitative profiling of proteins, phosphorylation, proteolysis, and histone modifications in solid and liquid biopsies from a range of pediatric cancers including leukemias, neuroblastomas, sarcomas, and high-risk brain tumors. With particular challenges of pediatric oncology in mind, some of the areas we investigate include:
- We are working towards real-time proteomic profiling of patient biopsies to stratify treatment recommendations in PROFYLE.
- In collaboration with our partners in BRAvE we study drug response and resistance in patient-derived cell, chicken chorioallantoic membrane (CAM) mouse models. These pre-clinical studies will help to increase access to cancer therapies.
- We study the use of proteomics to classify solid tumors that can not be differentiated based on histology, cytogenetics, or genomics.
- We investigate how will different patient-derived model systems (2D culture, mouse, CAM, zebrafish) recapitulate the proteome and PTM landscapes of primary patient cells.
- To enable relevant, age-matched comparisons we are heavily invested in gaining a better understanding of ‘normal’ ranges for protein abundance and protein modification in children as well as adolescents and young adults (AYA).
- We are working towards translation into clinical labs by developing biomarker panels and pediatric cancer-specific mass spectrometry assays.
Cell surface proteomics & new targets for cancer therapy
Pediatric malignancies have a far lower mutation and neoantigen rate than adult cancer. This renders many of the established approaches, that worked well for adult cancers, difficult. We have identified a new class of neoantigens that shows great potential in pediatric cancers.
In this project, we combine sensitive cell-surface proteomics of cancer models and patient biopsies with gene editing, engineering of recombinant proteins, and mouse studies. We evaluate the presence of neoantigens throughout progression and treatment and study the underlying mechanisms that lead to their formation. We also develop lead compounds that have the potential to specifically target the antigens and we develop diagnostic assays for companion diagnostics.
Cell-Cell Communication in the tumor microenvironment
Cells do not act in isolation. They secret a range of bioactive proteins that trigger a cellular response in neighboring and distant cells. We integrate cell biology, biochemistry, gene editing, bottom-up and top-down proteomics, and bioinformatics to study the role of post-translational modification in the regulation of select these extracellular signaling networks. Within our BRAvE platform, we recently established the chicken chorioallantoic membrane (CAM) model. This gives us an accessible and easy to manipulate system for mechanistic studies of findings we make in interstitial fluid of patient biopsies. Currently, we are particularly interested in understanding how secreted proteases, primarily Cathepsins and Matrix Metalloproteinases, remodel paracrine signaling in leukemia and neuroblastoma microenvironment.
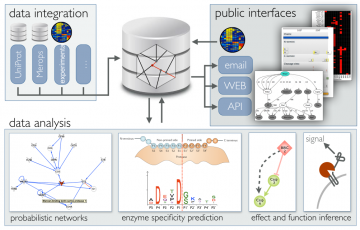
Proteoform function deconvolution
To harness the full diagnostic and treatment potential of proteoforms, we need new tools to better define and structure the proteoform landscape in an accessible and interpretable way and need to confirm that proteoform level measurements exceed protein level measurements. We develop algorithms for multi-scale data integration, proteoform function analysis, pattern recognition, and de-convolution of network effects in complex systems. Building on these we develop new biological knowledgebases and applications to improve the functional analysis of genomics and proteomics data and to guide personalized treatment decisions.
New technologies & methods in proteomics
Advances in the analysis of clinical specimens and new insights into complex disease biology are fundamentally enabled by breakthroughs in sample processing, protein biochemistry, and data analysis algorithms. We therefore continuously strive to improve existing- and develop new technologies (both, wet-lab and computational) for the analysis of complex biological specimens enabling a more comprehensive, specific, and sensitive investigation of smaller samples and more complex systems.